Abstract
Introduction
The clinical course and outcomes are heterogenous among patients (pts) with chronic lymphocytic leukemia (CLL). Somatic hypermutation (SHM) status of the IGHV gene (IGHV-SHM) is an established prognostic marker for pts receiving certain therapeutic regimens. Pts with mutated IGHV (IGHV-M) have a better prognosis vs pts with unmutated IGHV (IGHV-UM). We demonstrate that IGHV-SHM could be accurately assessed utilizing primers located downstream of CDR1.
Methods
Adaptive's clonoSEQ ® NGS-based assay identifies tumor specific sequence(s) using a set of multiplexed, locus-specific primers that include the Ig heavy chain variable gene (IGHV; starting at the end of CDR1), and the kappa and lambda light chain. Sequences are tagged and used to assess measurable residual disease (MRD).
SHM status is calculated as the percentage of somatic mutations deviating from the germline DNA sequence. IGHV-M was defined as ≥2% deviation. For patients with multiple IGHV sequences, the most mutated sequence determined IGHV-SHM status.
Pt samples from 5 studies (Table 1) were analyzed by clonoSEQ to determine IGHV-SHM status and compared to IGHV-SHM status determined by institutional standard laboratory (Sanger sequencing or institutional NGS). Four datasets were used from MD Anderson Cancer Center (MDACC). Set1 (n=31) was the positive control as eligibility criteria mandated inclusion of pts with IGHV-M status. Results were correlated with outcome data available from set2 (n=62). Pts with B-cell acute lymphoblastic leukemia served as a negative control (set3; n=246). Validation was performed with CLL samples (set4; n=21 and set5; n=49). IGHV-SHM analysis performed by Sanger sequencing (and NGS in a subset of set1 and set4) were the comparator for Adaptive's IGHV-SHM assessment.
Rearranged gDNA sequences from the IMGT database (est. 1989) were ascertained and IgBlast was used to annotate the domains and report the V-gene homology. To approximate a full-length (FL) rearrangement, 9080 sequences were identified where framework region 1 (FR1) had ≥ 75 nucleotides. We compared the mutation status obtained using this FL rearranged sequence to that same sequence beginning at the 5' end of FR2 (consistent with the 5' extent of the primers in the Adaptive Assay).
Results
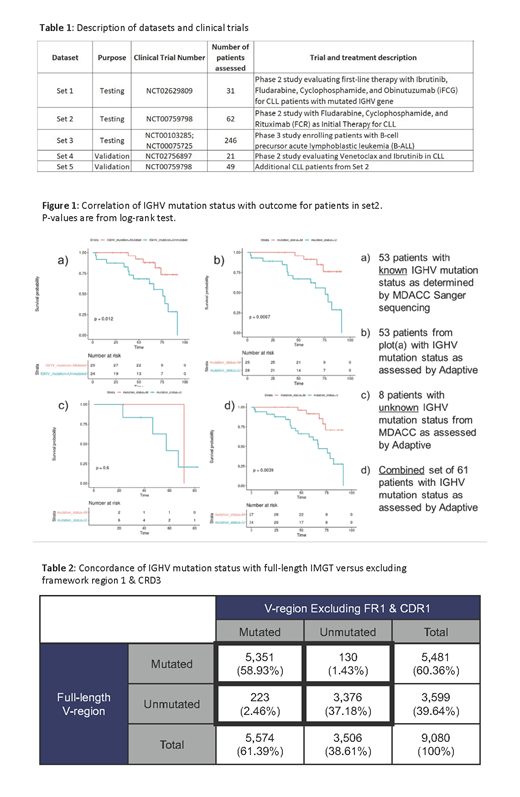
In set1, 30/31 pts were IGHV-M by the MDACC sequencing IGHV-SHM analysis. We obtained concordant IGHV-SHM status in 29/30 IGHV-M pts, and the only IGHV-UM pt. From set2, 29 pts with IGHV-M and 24 IGHV-UM were identified; 8 pts had unknown IGHV-SHM status. The IGHV-SHM Assay classified IGHV-M in 24/29 pts and IGHV-UM in 23/24 pts. In the negative control (set3), no IGHV-M pts were identified. In set2, stratifying PFS by Adaptive's IGHV-SHM status a p=0.0067 was non-inferior to the p=0.012 by Sanger sequencing (Figure 1 A-D). Inclusion of 8 pts with unknown IGHV-SHM status by Sanger sequencing yields a p=0.0039 (n=61). In the validation dataset (set4), IGHV-M was concordant for 4/4 IGHV-M pts and 13/14 IGHV-UM pts. In a separate validation dataset (set5), the Adaptive IGHV-SHM Assay was concordant for 16/17 IGHV-M pts and 25/28 IGHV-UM pts. 3 and 4 pts had unknown IGHV-SHM status by MDACC analysis in set4 and set5 respectively.
A concordance table compares mutation status with FL rearrangements from IMGT vs excluding FR1 & CDR1 (Table 2). 96.1% of the sequences had concordant SHM status. Of the 3.9% that are discordant, a majority are within ±1% of the 2% threshold, a recognized SHM "gray zone". Only 15 (0.17%) fall outside this borderline category and are categorized as IGHV-M converting to IGHV-UM based on the truncated analysis. Given improvements in sequencing since launching IMGT in 1989, it is likely that some sequencing quality issues may be present within the dataset such that the 0.17% discordance is an upper estimate of discordance.
Conclusions
135/147 CLL pts were concordant to independent assessments of IGHV-SHM status; IGHV-SHM status was assessed in additional 15 pts who could not be evaluated by MDACC comparator assay(s). The assessment of IGHV-SHM status correlated with PFS and standard determination of SHM. This analysis highlights the utility of the Adaptive assay to identify trackable sequences and assess IGHV-SHM status from the same baseline sample. The ability of a single platform to determine both SHM and enable subsequent MRD assessment streamlines the clinical management of pts with CLL.
Lee: Adaptive Biotechnologies: Current Employment. Ching: Adaptive Biotechnologies: Current Employment, Current equity holder in publicly-traded company. Howie: Adaptive Biotechnologies: Current Employment, Current equity holder in publicly-traded company, Divested equity in a private or publicly-traded company in the past 24 months. Thompson: Janssen: Consultancy, Honoraria; AbbVie: Other: Institution: Advisory/Consultancy, Honoraria, Research Grant/Funding; Gilead: Other: Institution: Advisory/Consultancy, Honoraria; Pharmacyclics: Other: Institution: Advisory/Consultancy, Honoraria, Research Grant/Funding; Adaptive Biotechnologies: Other: Institution: Advisory/Consultancy, Honoraria, Research Grant/Funding, Expert Testimony; Genentech: Other: Institution: Advisory/Consultancy, Honoraria, Research Grant/Funding; Amgen: Other: Institution: Honoraria, Research Grant/Funding. Jain: Beigene: Honoraria; Janssen: Honoraria; Bristol Myers Squibb: Honoraria, Research Funding; TG Therapeutics: Honoraria; Servier: Honoraria, Research Funding; AstraZeneca: Honoraria, Research Funding; ADC Therapeutics: Honoraria, Research Funding; Fate Therapeutics: Research Funding; Cellectis: Honoraria, Research Funding; Adaptive Biotechnologies: Honoraria, Research Funding; Incyte: Research Funding; Genentech: Honoraria, Research Funding; Pfizer: Research Funding; Aprea Therapeutics: Research Funding; Precision Biosciences: Honoraria, Research Funding; AbbVie: Honoraria, Research Funding; Pharmacyclics: Research Funding. Ferrajoli: BeiGene: Other: Advisory Board, Research Funding; Janssen: Other: Advisory Board ; AstraZeneca: Other: Advisory Board, Research Funding. Burger: TG Therapeutics: Other: Travel/Accommodations/Expenses, Research Funding, Speakers Bureau; AstraZeneca: Consultancy; Beigene: Research Funding, Speakers Bureau; Gilead: Consultancy, Other: Travel/Accommodations/Expenses, Research Funding, Speakers Bureau; Pharmacyclics LLC: Consultancy, Other: Travel/Accommodations/Expenses, Research Funding, Speakers Bureau; Novartis: Other: Travel/Accommodations/Expenses, Speakers Bureau; Janssen: Consultancy, Other: Travel/Accommodations/Expenses, Speakers Bureau. Wierda: Sunesis: Research Funding; Cyclacel: Research Funding; Miragen: Research Funding; Janssen: Research Funding; KITE Pharma: Research Funding; Karyopharm: Research Funding; Pharmacyclics LLC, an AbbVie Company: Research Funding; Gilead Sciences: Research Funding; Oncternal Therapeutics, Inc.: Research Funding; AstraZeneca: Research Funding; GSK/Novartis: Research Funding; Acerta Pharma Inc.: Research Funding; Xencor: Research Funding; Loxo Oncology, Inc.: Research Funding; Juno Therapeutics: Research Funding; Genentech: Research Funding; Genzyme Corporation: Consultancy; AbbVie: Research Funding. Kirsch: Adaptive Biotechnologies: Current Employment, Current equity holder in publicly-traded company.
Author notes
 This icon denotes a clinically relevant abstract
This icon denotes a clinically relevant abstract

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal